growth is a distinguishing feature of all living things. throughout the past century, the growth of living systems has fascinated physiologists, biologists, and clinical scientists alike. yet, most of their efforts remain exploratory and mainly qualitative. in the living matter lab, we use the fundamental laws of physics and create interactive simulation tools to predict the physiology and pathology of living systems. we strive to understand the mechanisms by which living systems grow, develop, evolve, and adapt. this is not always straightforward: living systems can undergo extreme deformations, change their mass, generate active contraction, and develop prestrain and residual stress. these phenomena are non-intuitive to traditional engineers and often difficult to grasp. they require us to advance the classical field theories of mechanics towards living matter physics, design our own mathematical models, and create our own simulation tools. in addition to a fundamental knowledge in biology and medicine, this demands a theoretical background in tensor calculus, continuum mechanics, and thermodynamics, paired with the ability to design efficient, stable, and robust computational tools.
An intriguing aspect of life is the interplay of form and function: Living systems manipulate their microenvironment and, conversely, the environment manipulates the living system. In my group, we explore these phenomena through iterative cycles of experimental testing, mathematical modeling, and computational simulation. Initially, we have focused exclusively on the cardiovascular system. We have established new methods to understand cardiomyopathy, mitral regurgitation, and heart failure. Using multiscale modeling, we have shown how the mechanisms of these pathologies propagate across the scales, from the molecular and cellular levels to the tissue and organ levels, to explain the loss of form and function. Our algorithms form the core of the Living Heart project, a multicenter initiative to create integrative, predictive models of the living human heart. We are now translating our approach to skin, muscle, the intestine, the lung, and the brain. This has important clinical applications in tissue engineering, tendon tear, asthma, bronchitis, autism, and schizophrenia. Beyond applications in biology and medicine, understanding the origin of shape has broad applications in the natural sciences including surface growth in thin films, mineral growth in geology, and rock folding in tectonophysics, projects we are pursuing jointly with collaborators from other disciplines.
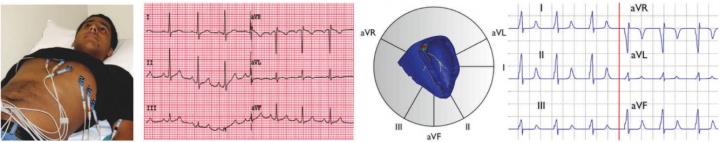
figure 1. validation of the new multi-scale simulation tool in terms of EKGs. in vivo acquired EKG recording based on a six-lead EKG (left), and in silico predicted EKG pattern based on a patient-specific whole heart model (right).
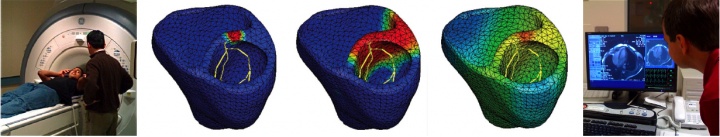
to calibrate and validate our model against clinical data, one of our students volunteered to have his EKG recording taken, see figure 1. to generate patient-specific anatomic models of the heart, we collaborate with professor michael mc connell in cardiovascular medicine. he took a series of MRI scans from which we generated a finite element mesh. figure 2 illustrates the time sequence of cardiac excitation simulated on the resulting patient-specific geometry. for the first time, we were able to generate an in silico EKG from the integration of all flux vectors projected onto six characteristic directions in space, see figure 1 [51]. we are excited about the incredible agreement between the in vivo recorded EKG, left, and the in silico predicted EKG, right. together with our collaborators in cardiac medicine, we plan to use our algorithms for both student education and new therapy design.
figure 2. patient-specific simulation of cardiac electrophysiology. MRI scan of human heart (left), simulated time sequence of cardiac excitation (middle), and diagnostic MRI images to generate anatomic human heart model (right).
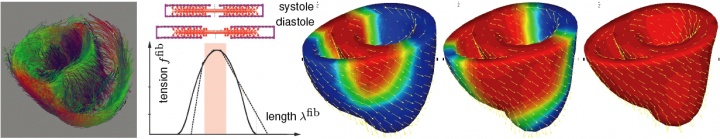
electric excitation induces an increased calcium concentration within the cell, which, in turn, initiates cellular contraction through actin-myosin filament sliding. to simulate these effects, existing simulation tools combine a finite-difference based excitation model with a finite-element based contraction model, having to pass information back and forth between both models at every instant in time. to reduce the risk of inherent instabilities associated with this explicit approach, existing simulation tools require an extremely fine spatial and temporal discretization. taking advantage of our core competence in multi-physics modeling, we have developed a fully coupled multi-field finite element approach to monolithically solve the electrical excitation problem and the mechanical equilibrium problem. this model can directly incorporate experimentally measured tension-length relations by mapping them onto patient-specific fiber orientations determined non-invasively through diffusion tensor MRI, see figure 3. for the first time, we have demonstrated a unified, fully coupled, extremely efficient, and robust solution technique that can reliably predict excitation-contraction patterns on single desktop computers with simulation times on the order of minutes.
figure 3. fiber orientation map from diffusion tensor MRI adopted from zhukov & barr [2003] (left), actin-myosin based filament sliding model with characteristic tension-length relation (middle), and simulation of excitation-contraction coupling in terms of bi-ventricular heart model with generic fiber orientation (right).
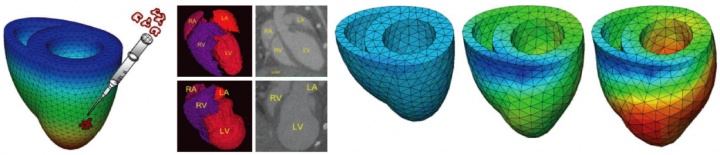
a leading cause of heart failure is myocardial infarction caused by the local death of heart muscle cells due to insufficient blood supply. as a result, the functional units of the myocardium, the cardiomyocytes, lose their contractile property and die. recently, stem cell therapy has emerged as a promising methodology for cardiac repair by injecting cells into the damaged tissue. in close collaboration with professors chris zarins in vascular surgery and sarah heilshorn in material sciences, we have developed computational tools to simulate the progression of infarct-induced heart failure to predict optimal cell injection sites. the underlying continuum model is based on the multiplicative decomposition of the deformation gradient into an elastic part and a growth part [25,54]. in the past, we have successfully applied this model in the context of open system thermodynamics [12,13,14,15] to simulate progressive plaque growth in arteries [34]. together with professor andreas menzel from the tu dortmund, germany [41], professor gerhard holzapfel from the tu graz, austria, and professors krishna garikipati and ellen arruda from the university of michigan, we have also developed algorithms to predict remodeling in arteries [36] and tendons [27,29]. we have now developed a new macroscopic growth and remodeling law for cardiac tissue motivated by microscopic changes in cardiomyocyte geometry that can be quantified experimentally through tissue histology. figure 4 shows our simulation of infarct-induced growth motivated by micro-ct images of mouse infarct models provided by our collaborator joe wu in radiology.
figure 4. virtual injection of stem-cell derived cardiomyocytes into the damaged myocardium (left). micro-ct images of infarct-induced growth in mouse model adopted from doyle et al. [2007] (middle). finite element simulation of infarct-induced growth (right).
recent studies in the wu lab have shown that stem cells may offer regenerative potential in infarct-induced heart failure. however, randomly injected cells lack the organization and physical structure required to create a cohesive, uniform tissue with continuous, synchronized beating. through a multidisciplinary collaboration with professors joe wu in radiology, chris zarins in vascular surgery, sarah heilshorn in material sciences, and beth pruitt in mechanical engineering, we are investigating the potential to engineer actively contracting stem-cell based tissue grafts to re-establish cardiac contraction and prevent heart failure. figure 5 displays our multi-scale finite element model of actively contracting cardiomyocytes seeded on a polymeric base layer derived in a collaboration with professor markus boel from the tu braunschweig, germany [48]. the in silico designed tissue constructs were calibrated, validated, and verified against in vitro engineered muscular thin films cultured by our collaborator professor kit parker at harvard university. our model is used to optimize the functional integration of a multi-layered tissue patch upon its implantation onto the ventricular wall.
figure 5. virtual implantation of tissue engineered vascular grafts onto the damaged myocardium (left). actively contracting cardiomyocytes seeded on polymeric base layer, experiment (top) vs. finite element simulation (bottom).
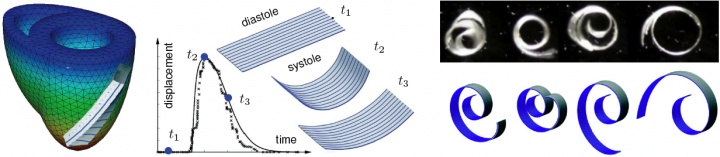
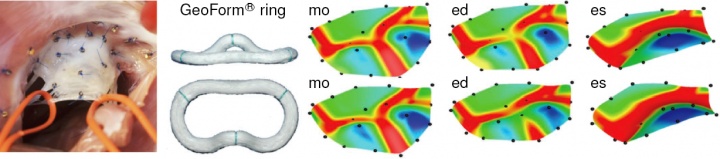
mitral regurgitation is a progressive, valvular disorder affecting 4 million people in the united states alone. the most common surgical approach to repair a leaking valve is to bring the leaflets closer together using annuloplasty rings. annuloplasty rings come in different shapes, materials, and sizes. choosing the optimal ring type largely depends on the surgeon's experience and personal preference. it is hypothesized that annuloplasty rings influence leaflet curvature, which in turn may considerably impact repair durability. in an attempt to guide the optimal ring choice and improve repair devices, we have quantified leaflet curvature in beating ovine hearts using in vivo acquired videofluoroscopic marker data provided by our collaborator professor craig miller in cardiothoracic surgery [56]. figure 6 documents the acute impact of annuloplasty ring implantation on the maximum principal leaflet curvature determined with our custom-made subdivision surface algorithm. using the same data set in an inverse finite element analysis has allowed us, for the first time, to identify the material parameters of the mitral valve in the beating heart [46]. a co-advised student in the lab of professor craig miller and professor neil ingels from the palo alto medical foundation identified the in vivo stiffness to be an order of magnitude larger than the in vitro measured tissue stiffness [50,52].
figure 6. intraoperative photograph showing 23 tantalium markers sewn on the mitral valve leaflet (left), annuloplasty ring (middle), and maximum principal curvature distribution in the leaflet with ring (top) and without ring (bottom).
our vision is to revolutionize thinking in regenerative medicine and induce a paradigm shift from empirical to predictive therapy design using multi-scale, multi-physics models derived from first principles.
previous research accomplishments 02
previous research accomplishments 01