| (44 intermediate revisions by one user not shown) | |||
| Line 1: | Line 1: | ||
| − | <div class="publicationheader">mission | + | <div class="publicationheader">check out our latest COVID-19 outbreak predictions... </div> |
| + | |||
| + | college campuses are COVID-19 superspreaders [https://www.forbes.com/sites/mishagajewski/2021/01/13/college-campuses-are-covid-19-superspreaders-study-says/?sh=2578217052fd forbes], [https://www.usnews.com/news/health-news/articles/2021-01-13/college-campuses-are-covid-superspreaders-study-finds US news & world report]<br> | ||
| + | are colleges superspreaders [https://www.insidehighered.com/news/2021/01/13/college-openings-led-increase-community-cases-research-says inside higher ed]<br> | ||
| + | college campuses can be superspreaders? [https://academictimes.com/college-campuses-can-be-superspreaders-but-safe-reopening-possible-study/ academic times]<br> | ||
| + | are college campuses COVID-19 superspreaders? [https://newsroom.taylorandfrancisgroup.com/new-study-suggests-that-college-campuses-are-covid-19-superspreaders/ t&f press release], [https://www.upi.com/Health_News/2021/01/13/College-campuses-may-be-COVID-19-super-spreaders-study-finds/5661610548574/ united press international]<br> | ||
| + | new study suggests that campuses are COVID-19 superspreaders [https://www.eurekalert.org/pub_releases/2021-01/tfg-nss011121.php eurekalert!], [https://consumer.healthday.com/b-1-13-study-confirms-college-campuses-as-super-spreader-centers-2649875013.html healthday]<br> | ||
| + | les campus universitaires sont des super-epandeurs [https://www.crumpi.com/2021/01/13/les-campus-universitaires-sont-des-super-epandeurs-de-covid-19-les-epidemies-sur-les-campus-locaux-se-propagent-rapidement-dans-tout-le-comte/ crumpi]<br> | ||
| + | reopening college campuses could initiate superspreading [https://www.news-medical.net/news/20201224/Reopening-college-campuses-could-initiate-COVID-19-superspreading.aspx med life sciences]<br> | ||
| + | a new class explores how to safely reopen a campus during covid-19 [https://engineering.stanford.edu/magazine/article/new-class-explores-how-safely-reopen-campus-during-covid-19 stanford report] [http://biomechanics.stanford.edu/paper/CMECH20b.pdf reopening paper]<br> | ||
| + | data-driven modeling of covid-19-lessons learned | ||
| + | [http://biomechanics.stanford.edu/paper/EML081220.jpg webinar announcement] | ||
| + | [https://www.youtube.com/watch?v=lBMPfltcDqs&feature=youtu.be webinar] | ||
| + | [http://biomechanics.stanford.edu/paper/EML20.pdf paper] <br> | ||
| + | how COVID-19 spread has been contained by travel bans [https://www.sciencedaily.com/releases/2020/05/200504191018.htm science daily] [https://infosurhoy.com/news-summary/how-covid-19-spread-has-been-contained-by-travel-bans/ infosurhoy] [http://biomechanics.stanford.edu/paper/CMBBE20.pdf cmbbe paper] <br> | ||
| + | covid-19 travel restrictions have saved millions of people [https://www.health24.com/Medical/Infectious-diseases/Coronavirus/covid-19-travel-restrictions-have-saved-millions-of-people-from-being-infected-mathematical-modelling-shows-20200507-2 health24 ] [https://medrxiv.org/cgi/content/short/2020.05.01.20088047v1 medRxiv Rvalues]<br> | ||
| + | how COVID-19 spread has been contained by travel bans [https://newsroom.taylorandfrancisgroup.com/how-covid-19-spread-has-been-contained-by-travel-bans/# press release] [http://biomechanics.stanford.edu/paper/CMBBE20.pdf cmbbe paper] [http://biomechanics.stanford.edu/paper/CMECH20a.pdf mobility paper]<br> | ||
| + | was haben die grenzschliessungen tatsachlich gebracht? [https://www.srf.ch/news/schweiz/kampf-gegen-das-coronavirus-was-haben-die-grenzschliessungen-tatsaechlich-gebracht swiss public radio] <br> | ||
| + | wie sinnvoll sind reisebeschrankungen wirklich? [https://www.welt.de/vermischtes/article207810451/Corona-Krise-und-Grenzen-Wie-sinnvoll-sind-Reisebeschraenkungen-wirklich.html welt] [http://biomechanics.stanford.edu/paper/CMBBE20.pdf cmbbe paper] <br> | ||
| + | so bremsen reisebeschrankungen das coronavirus aus [https://www.mdr.de/wissen/corona-reise-verbot-eindaemmung-covid-modell-100.html mdr wissen] [https://medrxiv.org/cgi/content/short/2020.05.01.20088047v1 medRxiv Rvalues]<br> | ||
| + | using math to understand COVID-19 outbreak dynamics [https://med.stanford.edu/cvi/mission/news_center/articles_announcements/using-math-to-understand-covid-19-outbreak-dynamics.html stanford medicine] [http://biomechanics.stanford.edu/paper/BMMB20.pdf bmmb paper] | ||
| + | [https://www.medrxiv.org/content/10.1101/2020.05.23.20111419v1 asymptomatic]<br> | ||
| + | predict covid's spread and recovery [https://engineering.stanford.edu/magazine/article/brain-inspired-model-can-help-predict-covid-s-spread-and-recovery?sf120699572=1 stanford engineering] [https://www.medrxiv.org/content/10.1101/2020.04.06.20055863v2 medRxiv in the us] [https://medrxiv.org/cgi/content/short/2020.04.18.20071035v1 medRxiv in europe]<br> | ||
| + | |||
| + | [[Image:research_us.jpg|720px]] | ||
| + | |||
| + | <div class="publicationheader">... and the other cool stuff we do</div> | ||
growth is a distinguishing feature of all living things. throughout the past century, the growth of living systems has fascinated physiologists, biologists, and clinical scientists alike. yet, most of their efforts remain exploratory and mainly qualitative. | growth is a distinguishing feature of all living things. throughout the past century, the growth of living systems has fascinated physiologists, biologists, and clinical scientists alike. yet, most of their efforts remain exploratory and mainly qualitative. | ||
| − | <b>in the living matter lab, we use the fundamental laws of physics and create interactive simulation tools to predict the physiology and pathology of living systems.</b> we strive to understand the mechanisms by which living systems grow, develop, evolve, and adapt. this is not always straightforward: living systems can undergo extreme deformations, change their mass, generate active contraction, and develop prestrain and residual stress. these phenomena are non-intuitive to traditional engineers and often difficult to grasp. they require us to advance the classical field theories of mechanics towards living matter physics, design our own mathematical models, and create our own simulation tools. | + | <b>in the living matter lab, we use the fundamental laws of physics and create interactive simulation tools to predict the physiology and pathology of living systems.</b> we strive to understand the mechanisms by which living systems grow, develop, evolve, and adapt. this is not always straightforward: living systems can undergo extreme deformations, change their mass, generate active contraction, and develop prestrain and residual stress. these phenomena are non-intuitive to traditional engineers and often difficult to grasp. they require us to advance the classical field theories of mechanics towards living matter physics, design our own mathematical models, and create our own simulation tools. here are some examples of projects that we are currently addressing: |
<div class="publicationheader">the living heart project</div> | <div class="publicationheader">the living heart project</div> | ||
| − | our heart is not only our most vital, but also our most complex organ: precisely controlled by the interplay of electrical and mechanical fields, it consists of four chambers and four valves, which act in concert to regulate its filling, ejection, and overall pump function. while numerous computational models exist to study either the electrical or the mechanical response of its individual chambers, the integrative electro-mechanical response of the whole heart remains poorly understood. with support of the nsf career award, our lab has designed the first fully coupled electro-mechanical model for excitation-contraction coupling within a single, unified finite element framework. our heart model has become a true success story: since may 2013, we have worked with the largest commercial finite element company, abaqus/dassault systemes, towards implementing our core algorithms into the abaqus infrastructure. together with the living heart team at dassault systemes, we have established the first four-chamber heart simulator for human heart function. the living heart project was officially launched in may 2014. since then, our work has received broad media attention and was featured in several scientific publications. to date, the living heart ecosystem has grown to 32 contributing member organizations, with more than 150 cardiovascular specialists from research, industry, and medicine, who have free access to our heart simulator to test and accelerate the development of the living heart via crowdsourcing. | + | our heart is not only our most vital, but also our most complex organ: precisely controlled by the interplay of electrical and mechanical fields, it consists of four chambers and four valves, which act in concert to regulate its filling, ejection, and overall pump function. while numerous computational models exist to study either the electrical or the mechanical response of its individual chambers, the integrative electro-mechanical response of the whole heart remains poorly understood. with support of the nsf career award, our lab has designed the first fully coupled electro-mechanical model for excitation-contraction coupling within a single, unified finite element framework. [http://biomechanics.stanford.edu/paper/LivingHeartEK.mp4 our heart model] has become a true success story: since may 2013, we have worked with the largest commercial finite element company, abaqus/dassault systemes, towards implementing our core algorithms into the abaqus infrastructure. together with the living heart team at dassault systemes, we have established the first four-chamber heart simulator for human heart function. the [http://www.3ds.com/heart living heart project] was officially launched in may 2014. since then, our work has received broad media attention and was featured in several |
| + | [http://www.sciencedirect.com/science/article/pii/S0997753814000564 scientific publications]. to date, the living heart ecosystem has grown to 32 contributing member organizations, with more than 150 cardiovascular specialists from research, industry, and medicine, who have free access to our heart simulator to test and accelerate the development of the living heart via crowdsourcing. | ||
[[Image:research01.jpg|720px]] | [[Image:research01.jpg|720px]] | ||
| Line 12: | Line 39: | ||
<b>figure 1.</b> the living heart project. in collaboration with abaqus/dassault systemes, we have created the first fully finite-element based whole heart model for realistic human heart simulations of excitation-contraction coupling. | <b>figure 1.</b> the living heart project. in collaboration with abaqus/dassault systemes, we have created the first fully finite-element based whole heart model for realistic human heart simulations of excitation-contraction coupling. | ||
| − | figure 1 illustrates our first prototype of the living heart model, which allows us to explore and understand the key features, physics, and technologies required to create an integrative, predictive model of the living human heart. ultimately, the living heart project will unite leading cardiovascular researchers, medical device manufacturers, regulatory agencies, and practicing cardiologists on the shared mission to develop and validate personalized digital human heart models and establish a unified foundation for in silico cardiovascular medicine | + | figure 1 illustrates our first prototype of the living heart model, which allows us to explore and understand the key features, physics, and technologies required to create an integrative, predictive model of the living human heart. ultimately, the living heart project will unite leading cardiovascular researchers, medical device manufacturers, regulatory agencies, and practicing cardiologists on the shared mission to develop and validate personalized digital human heart models and establish a unified foundation for in silico cardiovascular medicine. our computational models, which form the core of these efforts are documented in more than 20 peer-reviewed journal publications and in several [http://biomechanics.stanford.edu/paper/MEheart.pdf featured articles] and |
| − | + | [https://www.youtube.com/watch?v=6hsx_da1vVs animations]. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | [ | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | < | + | <div class="publicationheader">cardiac optogenetics</div> |
| − | + | ||
| − | + | heart disease is the primary cause of death in industrialized nations, claiming more than 16 million lives worldwide every year. in the united states alone, almost half a million people die each year as a result of heart rhythm disorders. despite its invasive nature, electrical stimulation remains the gold standard treatment to control rhythm disturbances through direct contact stimulation. together with our clinical collaborators, we have pioneered a novel technology to control heart rhythm disorders at a distance by means of light using a new concept known as [http://biomechanics.stanford.edu/paper/BIOPJ11.pdf optogenetics]. we have created mathematical models and computational tools to decipher the mechanisms by which optogenetics can regulate cardiac function with the ultimate goal to create a biological pacemaker of genetically engineered cardiac cells. | |
| − | + | [[File:research02.jpg|720px]] | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | <b>figure 2.</b> cardiac optogenetics. optical stimulation opens a cation channel to initiate a photocurrent, which increases the sodium concentration. this triggers an increase in the electric potential and initiates mechanical contraction. | |
| − | + | we prototyped a [http://biomechanics.stanford.edu/paper/JMPS12.pdf multiscale computational model] for the optogenetic control of human hearts, which we have calibrated across four biological scales as illustrated in figure 2. we used this model to make patient-specific predictions of ion channel dynamics, virtually probe landscapes of process parameters, and identify optimal photostimulation sequences in silico before testing them in vivo. this work is documented in five journal articles and has initiated a broad interest in the general media including several | |
| − | + | [http://www.nsf.gov/discoveries/disc_summ.jsp?cntn_id=129057 featured stories], | |
| + | [http://www.scienceupdate.com/?powerpress_pinw=8660-podcast podcasts], and | ||
| + | [http://biomechanics.stanford.edu/paper/Deutschlandfunk.mp3 radio interviews]. | ||
| − | <div class="publicationheader"> | + | <div class="publicationheader">characterizing living skin</div> |
| − | + | skin is our interface with the outside world. in its natural environment, it displays unique mechanical characteristics including prestrain and growth. while there is a general agreement on the physiological importance of these features, they remain poorly characterized mainly because they are difficult to access with standard laboratory techniques. our lab has established a novel, inexpensive technique to [http://biomechanics.stanford.edu/paper/ABM14.pdf characterize living skin] using multi-view stereo and isogeometric analysis. based on easy-to-create hand-held camera images, we quantify prestretch, deformation, and growth in a controlled porcine model of chronic skin expansion. over a period of five weeks, we gradually inflate subcutaneouly implanted balloons, take weekly photographs of the experimental scene, reconstruct the geometry from a tattooed surface grid, and create parametric representations of the skin surface. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | [[Image: | + | [[Image:research04.jpg|720px]] |
| − | <b>figure | + | <b>figure 3.</b> skin expansion in pediatric forehead reconstruction. |
| − | + | ||
| − | + | ||
| − | + | we analyzed these representations and quantified the average area prestretch to 1.44 and the average area growth to 2.25. we used these data to calibrate our skin growth model to simulate clinical cases of skin expansion in [http://biomechanics.stanford.edu/paper/JTBIO12.pdf pediatric forehead reconstruction]. our simulations accurately predict the clinically observed mechanical and structural response of living skin both acutely and chronically. our living skin model can easily be generalized to arbitrary biological membranes and serve as a valuable tool to virtually manipulate living systems, simply by means of changes in their mechanical environment. our skin research was initially motivated by a [http://biomechanics.stanford.edu/paper/JMPS11a.pdf class project] in fall 2010 and has, to date, inspired twelve conference papers and eleven [http://biomechanics.stanford.edu/paper/JTBIO12.pdf peer-reviewed journal articles]. | |
| − | + | <div class="publicationheader">physical biology of brain development</div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | the developing human brain remains one of the few unsolved mysteries of science. advancements in developmental biology, neuroscience, and medical imaging have brought us closer than ever to understand brain development in health and disease. however, the precise role of mechanics throughout this process remains under appreciated and poorly understood. we have recently shown that mechanical stretch plays a crucial role in [http://biomechanics.stanford.edu/paper/SREP14.pdf brain development]. | ||
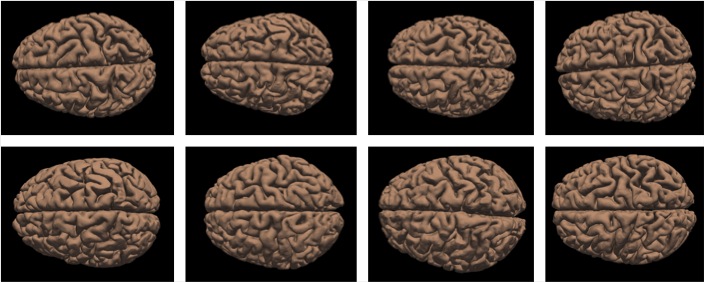
| − | [[Image: | + | [[Image:research03.jpg|720px]] |
| − | <b>figure | + | <b>figure 4.</b> variety of mammalian brains. caitlin, adrian, ellen, james, moritz, mona, rijk, maria. |
| − | + | ||
| − | + | using the nonlinear field theories of mechanics, supplemented by the theory of finite growth, we have modeled the human brain as a living system with a morphogenetically growing gray matter surface and a growing white matter core. this approach seamlessly integrates the two popular but competing hypotheses for cortical folding: axonal tension and differential growth. to characterize the stiffness of gray and white matter tissue, we have designed new protocols for [http://biomechanics.stanford.edu/paper/JMBBM15.pdf indentation testing] and demonstrated that white matter, with an average modulus of 1.9kpa, is stiffer than gray matter, with an average modulus of 1.4kpa. our simulations suggest that anisotropic white matter growth, as an emergent property from [http://biomechanics.stanford.edu/paper/ABME15a.pdf chronic axon elongation], intrinsically induces symmetry breaking, and predicts surface morphologies in agreement with magnetic resonance images from very preterm neonates. our model predicts that deviations in cortical thickness, elasticity, and growth induce morphological abnormalities. using the gyrification index, the ratio between the total and exposed surface area, we have shown that these abnormalities agree with the classical pathologies of lissencephaly and polymicrogyria. understanding the mechanisms of human brain development has direct implications on the diagnostics and treatment of neurological disorders, including epilepsy, schizophrenia, and autism. In the past year, we have published six peer reviewed | |
| − | in | + | [http://biomechanics.stanford.edu/paper/JMPS14b.pdf journal articles] and edited a |
| − | + | [http://link.springer.com/article/10.1007/s10237-015-0662-4 multi-author review] on brain mechanics. | |
[[previous research accomplishments 02]]<br> | [[previous research accomplishments 02]]<br> | ||
[[previous research accomplishments 01]]<br> | [[previous research accomplishments 01]]<br> | ||
Latest revision as of 13:38, 13 January 2021
college campuses are COVID-19 superspreaders forbes, US news & world report
are colleges superspreaders inside higher ed
college campuses can be superspreaders? academic times
are college campuses COVID-19 superspreaders? t&f press release, united press international
new study suggests that campuses are COVID-19 superspreaders eurekalert!, healthday
les campus universitaires sont des super-epandeurs crumpi
reopening college campuses could initiate superspreading med life sciences
a new class explores how to safely reopen a campus during covid-19 stanford report reopening paper
data-driven modeling of covid-19-lessons learned
webinar announcement
webinar
paper
how COVID-19 spread has been contained by travel bans science daily infosurhoy cmbbe paper
covid-19 travel restrictions have saved millions of people health24 medRxiv Rvalues
how COVID-19 spread has been contained by travel bans press release cmbbe paper mobility paper
was haben die grenzschliessungen tatsachlich gebracht? swiss public radio
wie sinnvoll sind reisebeschrankungen wirklich? welt cmbbe paper
so bremsen reisebeschrankungen das coronavirus aus mdr wissen medRxiv Rvalues
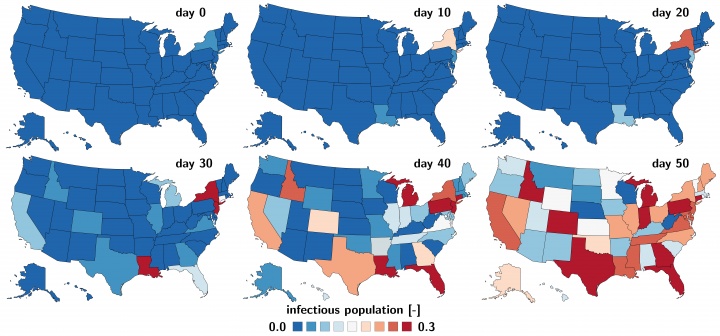
using math to understand COVID-19 outbreak dynamics stanford medicine bmmb paper
asymptomatic
predict covid's spread and recovery stanford engineering medRxiv in the us medRxiv in europe
growth is a distinguishing feature of all living things. throughout the past century, the growth of living systems has fascinated physiologists, biologists, and clinical scientists alike. yet, most of their efforts remain exploratory and mainly qualitative. in the living matter lab, we use the fundamental laws of physics and create interactive simulation tools to predict the physiology and pathology of living systems. we strive to understand the mechanisms by which living systems grow, develop, evolve, and adapt. this is not always straightforward: living systems can undergo extreme deformations, change their mass, generate active contraction, and develop prestrain and residual stress. these phenomena are non-intuitive to traditional engineers and often difficult to grasp. they require us to advance the classical field theories of mechanics towards living matter physics, design our own mathematical models, and create our own simulation tools. here are some examples of projects that we are currently addressing:
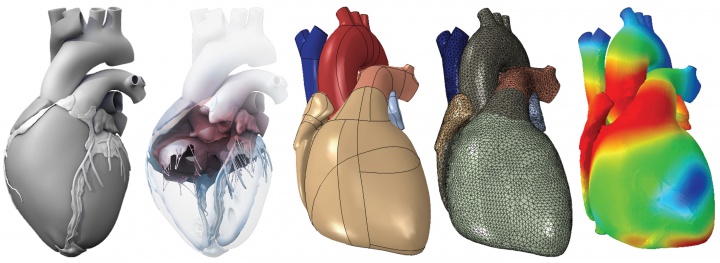
our heart is not only our most vital, but also our most complex organ: precisely controlled by the interplay of electrical and mechanical fields, it consists of four chambers and four valves, which act in concert to regulate its filling, ejection, and overall pump function. while numerous computational models exist to study either the electrical or the mechanical response of its individual chambers, the integrative electro-mechanical response of the whole heart remains poorly understood. with support of the nsf career award, our lab has designed the first fully coupled electro-mechanical model for excitation-contraction coupling within a single, unified finite element framework. our heart model has become a true success story: since may 2013, we have worked with the largest commercial finite element company, abaqus/dassault systemes, towards implementing our core algorithms into the abaqus infrastructure. together with the living heart team at dassault systemes, we have established the first four-chamber heart simulator for human heart function. the living heart project was officially launched in may 2014. since then, our work has received broad media attention and was featured in several scientific publications. to date, the living heart ecosystem has grown to 32 contributing member organizations, with more than 150 cardiovascular specialists from research, industry, and medicine, who have free access to our heart simulator to test and accelerate the development of the living heart via crowdsourcing.
figure 1. the living heart project. in collaboration with abaqus/dassault systemes, we have created the first fully finite-element based whole heart model for realistic human heart simulations of excitation-contraction coupling.
figure 1 illustrates our first prototype of the living heart model, which allows us to explore and understand the key features, physics, and technologies required to create an integrative, predictive model of the living human heart. ultimately, the living heart project will unite leading cardiovascular researchers, medical device manufacturers, regulatory agencies, and practicing cardiologists on the shared mission to develop and validate personalized digital human heart models and establish a unified foundation for in silico cardiovascular medicine. our computational models, which form the core of these efforts are documented in more than 20 peer-reviewed journal publications and in several featured articles and animations.
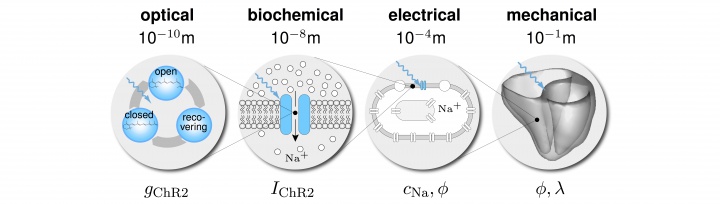
heart disease is the primary cause of death in industrialized nations, claiming more than 16 million lives worldwide every year. in the united states alone, almost half a million people die each year as a result of heart rhythm disorders. despite its invasive nature, electrical stimulation remains the gold standard treatment to control rhythm disturbances through direct contact stimulation. together with our clinical collaborators, we have pioneered a novel technology to control heart rhythm disorders at a distance by means of light using a new concept known as optogenetics. we have created mathematical models and computational tools to decipher the mechanisms by which optogenetics can regulate cardiac function with the ultimate goal to create a biological pacemaker of genetically engineered cardiac cells.
figure 2. cardiac optogenetics. optical stimulation opens a cation channel to initiate a photocurrent, which increases the sodium concentration. this triggers an increase in the electric potential and initiates mechanical contraction.
we prototyped a multiscale computational model for the optogenetic control of human hearts, which we have calibrated across four biological scales as illustrated in figure 2. we used this model to make patient-specific predictions of ion channel dynamics, virtually probe landscapes of process parameters, and identify optimal photostimulation sequences in silico before testing them in vivo. this work is documented in five journal articles and has initiated a broad interest in the general media including several featured stories, podcasts, and radio interviews.
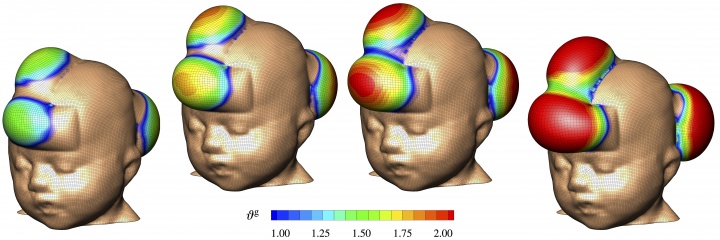
skin is our interface with the outside world. in its natural environment, it displays unique mechanical characteristics including prestrain and growth. while there is a general agreement on the physiological importance of these features, they remain poorly characterized mainly because they are difficult to access with standard laboratory techniques. our lab has established a novel, inexpensive technique to characterize living skin using multi-view stereo and isogeometric analysis. based on easy-to-create hand-held camera images, we quantify prestretch, deformation, and growth in a controlled porcine model of chronic skin expansion. over a period of five weeks, we gradually inflate subcutaneouly implanted balloons, take weekly photographs of the experimental scene, reconstruct the geometry from a tattooed surface grid, and create parametric representations of the skin surface.
figure 3. skin expansion in pediatric forehead reconstruction.
we analyzed these representations and quantified the average area prestretch to 1.44 and the average area growth to 2.25. we used these data to calibrate our skin growth model to simulate clinical cases of skin expansion in pediatric forehead reconstruction. our simulations accurately predict the clinically observed mechanical and structural response of living skin both acutely and chronically. our living skin model can easily be generalized to arbitrary biological membranes and serve as a valuable tool to virtually manipulate living systems, simply by means of changes in their mechanical environment. our skin research was initially motivated by a class project in fall 2010 and has, to date, inspired twelve conference papers and eleven peer-reviewed journal articles.
the developing human brain remains one of the few unsolved mysteries of science. advancements in developmental biology, neuroscience, and medical imaging have brought us closer than ever to understand brain development in health and disease. however, the precise role of mechanics throughout this process remains under appreciated and poorly understood. we have recently shown that mechanical stretch plays a crucial role in brain development.
figure 4. variety of mammalian brains. caitlin, adrian, ellen, james, moritz, mona, rijk, maria.
using the nonlinear field theories of mechanics, supplemented by the theory of finite growth, we have modeled the human brain as a living system with a morphogenetically growing gray matter surface and a growing white matter core. this approach seamlessly integrates the two popular but competing hypotheses for cortical folding: axonal tension and differential growth. to characterize the stiffness of gray and white matter tissue, we have designed new protocols for indentation testing and demonstrated that white matter, with an average modulus of 1.9kpa, is stiffer than gray matter, with an average modulus of 1.4kpa. our simulations suggest that anisotropic white matter growth, as an emergent property from chronic axon elongation, intrinsically induces symmetry breaking, and predicts surface morphologies in agreement with magnetic resonance images from very preterm neonates. our model predicts that deviations in cortical thickness, elasticity, and growth induce morphological abnormalities. using the gyrification index, the ratio between the total and exposed surface area, we have shown that these abnormalities agree with the classical pathologies of lissencephaly and polymicrogyria. understanding the mechanisms of human brain development has direct implications on the diagnostics and treatment of neurological disorders, including epilepsy, schizophrenia, and autism. In the past year, we have published six peer reviewed journal articles and edited a multi-author review on brain mechanics.
previous research accomplishments 02
previous research accomplishments 01