(→additional reading) |
|||
| Line 153: | Line 153: | ||
==additional reading== | ==additional reading== | ||
| − | + | bar-on et al., sars-cov-2 (covid-19) by the numbers, elife 9 (2020) e57309. | |
| − | [http://biomechanics.stanford.edu/ | + | [http://biomechanics.stanford.edu/me233_20/reading/baron20.pdf (download)]<br> |
| − | + | bauer f, compartment models in epidemiology, mathematical epidemiology (2008) 19-79. | |
| − | + | [http://biomechanics.stanford.edu/me233_20/reading/bauer20.pdf (download)]<br> | |
| − | [http:// | + | |
| − | + | hethcode hw, the mathematics of infectious disease, siam review 42 (2020) 599-653. | |
| − | + | [http://biomechanics.stanford.edu/me233_20/reading/hethcote20.pdf (download)]<br> | |
| − | + | ||
| − | [http://biomechanics.stanford.edu/ | + | |
| − | + | dietz k, the estimation of the basic reproduction number for infectious diseases, stat meth med res 2 (1993) 23-41. | |
| − | + | [http://biomechanics.stanford.edu/me233_20/reading/dietz93.pdf (download)]<br> | |
| − | + | ||
| − | [ | + | |
| − | + | peirlinck m, et al. outbreak dynamics of covid-19 in china and the united states. medRxiv: doi:10.1101/2020.04.06.20055863. | |
| − | [http://biomechanics.stanford.edu/ | + | [http://biomechanics.stanford.edu/me233_20/reading/peirlick20a.pdf (download)]<br> |
| − | + | park et al., reconciling early-outbreak estimates of the basic reproduction number and its uncertainty. j royal soc interface 17 (2020) 20200144. | |
| − | the | + | [http://biomechanics.stanford.edu/me233_20/reading/park20.pdf (download)]<br> |
| − | j | + | |
| − | [http://biomechanics.stanford.edu/ | + | |
| − | + | ioannis j, the invection fatality rate of covid-19 inferred from seroprevalence data, medRxiv, doi:10.1101/2020.05.13.20101253. | |
| − | + | [http://biomechanics.stanford.edu/me233_20/reading/ioannis20.pdf (download)]<br> | |
| − | + | ||
| − | [http://biomechanics.stanford.edu/ | + | |
| − | + | peirlinck m et al., visualizing the invisible: the effect of asymptomatic transmission, medRxiv, doi: 10.1101/2020.05.23.20111419 | |
| − | + | [http://biomechanics.stanford.edu/me233_20/reading/peirlinck20b.pdf (download)]<br> | |
| − | + | ||
| − | [ | + | |
| − | + | wilder-smith a, freedman do. isolation, quarantine, social distancing | |
| − | + | and community containment, j travel med (2020) 1-4. | |
| − | + | [http://biomechanics.stanford.edu/me233_20/reading/wilder20.pdf (download)]<br> | |
| − | [http://biomechanics.stanford.edu/ | + | |
| − | + | dehning et al., inferring change points in the spread of COVID-19 reveals the effectiveness of interventions, science doi:10.1126/science.abb9789. | |
| − | + | [http://biomechanics.stanford.edu/me233_20/reading/dehning20.pdf (download)]<br> | |
| − | + | ||
| − | [http://biomechanics.stanford.edu/ | + | |
| − | + | linka et al., the reproduction number of covid-19 and its correlation with public health interventions, comp mech, doi:10.1007/s00466-020-01880-8. | |
| − | + | [http://biomechanics.stanford.edu/me233_20/reading/linka20a.pdf (download)]<br> | |
| − | + | ||
| − | [http://biomechanics.stanford.edu/ | + | |
| − | + | pastor-satorras r et al., epidemic processes in complex networks, rev mod phys 87 (2015) 926-973. | |
| − | + | [http://biomechanics.stanford.edu/me233_20/reading/pastor20.pdf (download)]<br> | |
| − | + | ||
| − | [http://biomechanics.stanford.edu/ | + | |
| − | + | linka k et al. outbreak dynamics of covid-19 in europe and the effect of travel restrictions. comp meth biomech biomed eng; 2020; 23:710-717. | |
| − | + | [http://biomechanics.stanford.edu/me233_20/reading/linka20b.pdf (download)]<br> | |
| − | + | ||
| − | [http://biomechanics.stanford.edu/ | + | |
| − | + | linka k et al. global and local mobility as a barometer for COVID-19 dynamics. medRxiv doi:2020.06.13.20130658. | |
| − | + | [http://biomechanics.stanford.edu/me233_20/reading/linka20d.pdf (download)]<br> | |
| − | + | ||
| − | [http://biomechanics.stanford.edu/ | + | |
| − | + | linka k et al. is it safe to lift COVID-19 travel bans. the newfoundland story. comp mech doi:10.1007/s00466-020-01899-x. | |
| − | + | [http://biomechanics.stanford.edu/me233_20/reading/linka20c.pdf (download)]<br> | |
| − | + | ||
| − | [http://biomechanics.stanford.edu/ | + | |
| − | + | anderson rm, may rm, directly transmitted infectious diseases: control by vaccination, science 215 (1982) 1053-1060 | |
| − | + | [http://biomechanics.stanford.edu/me233_20/reading/anderson82.pdf (download)]<br> | |
| − | + | ||
| − | [http://biomechanics.stanford.edu/ | + | grassly nc, fraser c, seasonal infectious disease epidemiology, proc royal |
| + | soc b 273 (2006) 2541-2550. | ||
| + | [http://biomechanics.stanford.edu/me233_20/reading/grally06.pdf (download)]<br> | ||
| + | |||
| + | kuhl e. data-driven modeling of COVID-19 - lessons learned. extr mech lett; doi:10.1016/j.eml.2020.100921. | ||
| + | [http://biomechanics.stanford.edu/me233_20/reading/kuhl20.pdf (download)]<br> | ||
Revision as of 16:47, 20 August 2020
Contents |
fall 21 - me233 - data-driven modeling of covid-19
|
me 233 - data-driven modeling of covid-19 ellen kuhl
kevin linka fall 2021 |
dissecting brains

| 
| 
| 
|

| 
| 
| 
|

| 
| 
| 
|

| 
| 
| 
|
objectives
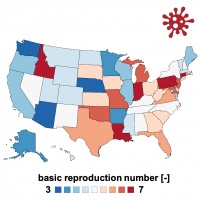
understanding the outbreak dynamics of COVID-19 through the lens of mathematical models is an elusive but significant goal. within only half a year, the COVID-19 pandemic has resulted in more than 20 million reported cases across 188 countries with more than 700,000 deaths worldwide. this has generated an unprecedented volume of data; yet, the precise role of mathematical modeling in providing quantitative insight into the COVID-19 pandemic remains a topic of ongoing debate. this course discusses how to design computational tools to understand the COVID-19 outbreak. we focus on mathematical epidemiology, infectious disease models, concepts of the effective reproduction number and herd immunity, network modeling, outbreak dynamics and outbreak control, bayesian methods, model calibration and validation, prediction and uncertainty quantification. we highlight the early success of classical models for infectious diseases and show why these models fail to predict the current outbreak dynamics of COVID-19. we illustrate how data-driven modeling can integrate classical epidemiology modeling and machine learning to infer critical disease parameters—in real time—from reported case data to make informed predictions and guide political decision making. We critically discuss questions that current models can and cannot answer and showcase controversial decisions around the early outbreak dynamics, outbreak control, and exit strategies.
grading
- 15 % homework 01 - mathematical epidemiology of covid-19
- 15 % homework 02 - early outbreak dynamics of covid-19
- 15 % homework 03 - outbreak control of covid-19
- 25 % final project - presentation graded by class
- 30 % final project - report graded by instructors
final project
harris tc, de rooij r, kuhl e.
the shrinking brain: cerebral atrophy following traumatic brain injury.
ann biomed eng. 2019; 47:1941-1959.
(download)
weickenmeier j, kurt m, ozkaya e, wintermark m, butts pauly k, kuhl e.
magnetic resonance elastography of the brain: a comparison between pigs and humans.
j mech beh biomed mat. 2018; 77:702-710.
(download)
wu lc, ye pp, kuo c, laksari k, camarillo d, kuhl e. pilot findings of brain displacements and deformations during roller coaster rides. j neurotrauma. 2017; 34:3198-3205. (download)
lejeune e, javili a, weickenmeier j, kuhl e, linder c. tri-layer wrinkling as a mechanism for anchoring center initiation in the developing cerebellum. soft matter. 2016;12:5613-5620. (download)
ploch cc, mansi cssa, jayamohan j, kuhl e. using 3D printing to create personalized brain models for neurosurgical training and preoperative planning. world neurosurg. 2016;90:668-674. (download), (perspectives)
syllabus
| day | date | topic | slides | homework | |
|---|---|---|---|---|---|
| tue | sept | 15 | introduction to covid-19 | s01 | |
| thu | sept | 17 | I. infectious disesaes - a brief history | s02 | |
| tue | sept | 22 | II. mathematical epidemiology - 1. intro to compartment modeling | s03 | |
| thu | setp | 24 | II. mathematical epidemiology - 2. compartment models | s04 | h01 |
| tue | sept | 29 | II. mathematical epidemiology - 3. endemic disease modeling | s05 | |
| thu | oct | 01 | III. data-driven modeling - 1. compartment modeling of covid-19 | s06 | |
| tue | oct | 06 | III. data-driven modeling - 2. early outbreak dynamics of covid-19 | s07 | |
| thu | oct | 08 | III. data-driven modeling - 3. asymptomatic transmission of covid-19 | s08 | h02 |
| tue | oct | 13 | III. data-driven modeling - 4. inferring outbreak dynamics of covid-19 | s09 | |
| thu | oct | 15 | IV. modeling outbreak control - 1. managing infectious diseases | s10 | |
| tue | oct | 20 | IV. modeling outbreak control - 2. change-point modeling of covid-19 | s11 | |
| thu | oct | 22 | IV. modeling outbreak control - 3. dynamic modeling of covid-19 | s12 | h03 |
| tue | oct | 27 | V. network modeling - 1. network modeling of epidemiology | s13 | |
| thu | oct | 29 | V. network modeling - 2. network modeling of covid-19 | s14 | |
| tue | nov | 03 | V. network modeling - 3. dynamic network modeling of covid-19 | s15 | |
| thu | oct | 05 | VI. informing political decision making - exit strategies | s16 | p01 |
| tue | oct | 10 | VI. informing political decision making - vaccination strategies | s17 | |
| thu | oct | 12 | VI. informing political decision making - the second wave | s18 | |
| tue | oct | 17 | data-driven modeling of covid-19 - lessons learned | s19 | |
| thu | oct | 19 | data-driven modeling of covid-19 - final project presentations | s20 |
python files
we'll share the python files for the projects here
additional reading
bar-on et al., sars-cov-2 (covid-19) by the numbers, elife 9 (2020) e57309.
(download)
bauer f, compartment models in epidemiology, mathematical epidemiology (2008) 19-79.
(download)
hethcode hw, the mathematics of infectious disease, siam review 42 (2020) 599-653.
(download)
dietz k, the estimation of the basic reproduction number for infectious diseases, stat meth med res 2 (1993) 23-41.
(download)
peirlinck m, et al. outbreak dynamics of covid-19 in china and the united states. medRxiv: doi:10.1101/2020.04.06.20055863.
(download)
park et al., reconciling early-outbreak estimates of the basic reproduction number and its uncertainty. j royal soc interface 17 (2020) 20200144.
(download)
ioannis j, the invection fatality rate of covid-19 inferred from seroprevalence data, medRxiv, doi:10.1101/2020.05.13.20101253.
(download)
peirlinck m et al., visualizing the invisible: the effect of asymptomatic transmission, medRxiv, doi: 10.1101/2020.05.23.20111419
(download)
wilder-smith a, freedman do. isolation, quarantine, social distancing
and community containment, j travel med (2020) 1-4.
(download)
dehning et al., inferring change points in the spread of COVID-19 reveals the effectiveness of interventions, science doi:10.1126/science.abb9789.
(download)
linka et al., the reproduction number of covid-19 and its correlation with public health interventions, comp mech, doi:10.1007/s00466-020-01880-8.
(download)
pastor-satorras r et al., epidemic processes in complex networks, rev mod phys 87 (2015) 926-973.
(download)
linka k et al. outbreak dynamics of covid-19 in europe and the effect of travel restrictions. comp meth biomech biomed eng; 2020; 23:710-717.
(download)
linka k et al. global and local mobility as a barometer for COVID-19 dynamics. medRxiv doi:2020.06.13.20130658.
(download)
linka k et al. is it safe to lift COVID-19 travel bans. the newfoundland story. comp mech doi:10.1007/s00466-020-01899-x.
(download)
anderson rm, may rm, directly transmitted infectious diseases: control by vaccination, science 215 (1982) 1053-1060
(download)
grassly nc, fraser c, seasonal infectious disease epidemiology, proc royal
soc b 273 (2006) 2541-2550.
(download)
kuhl e. data-driven modeling of COVID-19 - lessons learned. extr mech lett; doi:10.1016/j.eml.2020.100921.
(download)